Part 1
Contents
Part 1#
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "all"
x = 1
y = 2
x
y
1
2
Load sample dataset#
from sklearn.datasets import load_iris
from sklearn.datasets import load_diabetes
iris_dataset = load_iris()
iris_dataset
{'data': array([[5.1, 3.5, 1.4, 0.2],
[4.9, 3. , 1.4, 0.2],
[4.7, 3.2, 1.3, 0.2],
[4.6, 3.1, 1.5, 0.2],
[5. , 3.6, 1.4, 0.2],
[5.4, 3.9, 1.7, 0.4],
[4.6, 3.4, 1.4, 0.3],
[5. , 3.4, 1.5, 0.2],
[4.4, 2.9, 1.4, 0.2],
[4.9, 3.1, 1.5, 0.1],
[5.4, 3.7, 1.5, 0.2],
[4.8, 3.4, 1.6, 0.2],
[4.8, 3. , 1.4, 0.1],
[4.3, 3. , 1.1, 0.1],
[5.8, 4. , 1.2, 0.2],
[5.7, 4.4, 1.5, 0.4],
[5.4, 3.9, 1.3, 0.4],
[5.1, 3.5, 1.4, 0.3],
[5.7, 3.8, 1.7, 0.3],
[5.1, 3.8, 1.5, 0.3],
[5.4, 3.4, 1.7, 0.2],
[5.1, 3.7, 1.5, 0.4],
[4.6, 3.6, 1. , 0.2],
[5.1, 3.3, 1.7, 0.5],
[4.8, 3.4, 1.9, 0.2],
[5. , 3. , 1.6, 0.2],
[5. , 3.4, 1.6, 0.4],
[5.2, 3.5, 1.5, 0.2],
[5.2, 3.4, 1.4, 0.2],
[4.7, 3.2, 1.6, 0.2],
[4.8, 3.1, 1.6, 0.2],
[5.4, 3.4, 1.5, 0.4],
[5.2, 4.1, 1.5, 0.1],
[5.5, 4.2, 1.4, 0.2],
[4.9, 3.1, 1.5, 0.2],
[5. , 3.2, 1.2, 0.2],
[5.5, 3.5, 1.3, 0.2],
[4.9, 3.6, 1.4, 0.1],
[4.4, 3. , 1.3, 0.2],
[5.1, 3.4, 1.5, 0.2],
[5. , 3.5, 1.3, 0.3],
[4.5, 2.3, 1.3, 0.3],
[4.4, 3.2, 1.3, 0.2],
[5. , 3.5, 1.6, 0.6],
[5.1, 3.8, 1.9, 0.4],
[4.8, 3. , 1.4, 0.3],
[5.1, 3.8, 1.6, 0.2],
[4.6, 3.2, 1.4, 0.2],
[5.3, 3.7, 1.5, 0.2],
[5. , 3.3, 1.4, 0.2],
[7. , 3.2, 4.7, 1.4],
[6.4, 3.2, 4.5, 1.5],
[6.9, 3.1, 4.9, 1.5],
[5.5, 2.3, 4. , 1.3],
[6.5, 2.8, 4.6, 1.5],
[5.7, 2.8, 4.5, 1.3],
[6.3, 3.3, 4.7, 1.6],
[4.9, 2.4, 3.3, 1. ],
[6.6, 2.9, 4.6, 1.3],
[5.2, 2.7, 3.9, 1.4],
[5. , 2. , 3.5, 1. ],
[5.9, 3. , 4.2, 1.5],
[6. , 2.2, 4. , 1. ],
[6.1, 2.9, 4.7, 1.4],
[5.6, 2.9, 3.6, 1.3],
[6.7, 3.1, 4.4, 1.4],
[5.6, 3. , 4.5, 1.5],
[5.8, 2.7, 4.1, 1. ],
[6.2, 2.2, 4.5, 1.5],
[5.6, 2.5, 3.9, 1.1],
[5.9, 3.2, 4.8, 1.8],
[6.1, 2.8, 4. , 1.3],
[6.3, 2.5, 4.9, 1.5],
[6.1, 2.8, 4.7, 1.2],
[6.4, 2.9, 4.3, 1.3],
[6.6, 3. , 4.4, 1.4],
[6.8, 2.8, 4.8, 1.4],
[6.7, 3. , 5. , 1.7],
[6. , 2.9, 4.5, 1.5],
[5.7, 2.6, 3.5, 1. ],
[5.5, 2.4, 3.8, 1.1],
[5.5, 2.4, 3.7, 1. ],
[5.8, 2.7, 3.9, 1.2],
[6. , 2.7, 5.1, 1.6],
[5.4, 3. , 4.5, 1.5],
[6. , 3.4, 4.5, 1.6],
[6.7, 3.1, 4.7, 1.5],
[6.3, 2.3, 4.4, 1.3],
[5.6, 3. , 4.1, 1.3],
[5.5, 2.5, 4. , 1.3],
[5.5, 2.6, 4.4, 1.2],
[6.1, 3. , 4.6, 1.4],
[5.8, 2.6, 4. , 1.2],
[5. , 2.3, 3.3, 1. ],
[5.6, 2.7, 4.2, 1.3],
[5.7, 3. , 4.2, 1.2],
[5.7, 2.9, 4.2, 1.3],
[6.2, 2.9, 4.3, 1.3],
[5.1, 2.5, 3. , 1.1],
[5.7, 2.8, 4.1, 1.3],
[6.3, 3.3, 6. , 2.5],
[5.8, 2.7, 5.1, 1.9],
[7.1, 3. , 5.9, 2.1],
[6.3, 2.9, 5.6, 1.8],
[6.5, 3. , 5.8, 2.2],
[7.6, 3. , 6.6, 2.1],
[4.9, 2.5, 4.5, 1.7],
[7.3, 2.9, 6.3, 1.8],
[6.7, 2.5, 5.8, 1.8],
[7.2, 3.6, 6.1, 2.5],
[6.5, 3.2, 5.1, 2. ],
[6.4, 2.7, 5.3, 1.9],
[6.8, 3. , 5.5, 2.1],
[5.7, 2.5, 5. , 2. ],
[5.8, 2.8, 5.1, 2.4],
[6.4, 3.2, 5.3, 2.3],
[6.5, 3. , 5.5, 1.8],
[7.7, 3.8, 6.7, 2.2],
[7.7, 2.6, 6.9, 2.3],
[6. , 2.2, 5. , 1.5],
[6.9, 3.2, 5.7, 2.3],
[5.6, 2.8, 4.9, 2. ],
[7.7, 2.8, 6.7, 2. ],
[6.3, 2.7, 4.9, 1.8],
[6.7, 3.3, 5.7, 2.1],
[7.2, 3.2, 6. , 1.8],
[6.2, 2.8, 4.8, 1.8],
[6.1, 3. , 4.9, 1.8],
[6.4, 2.8, 5.6, 2.1],
[7.2, 3. , 5.8, 1.6],
[7.4, 2.8, 6.1, 1.9],
[7.9, 3.8, 6.4, 2. ],
[6.4, 2.8, 5.6, 2.2],
[6.3, 2.8, 5.1, 1.5],
[6.1, 2.6, 5.6, 1.4],
[7.7, 3. , 6.1, 2.3],
[6.3, 3.4, 5.6, 2.4],
[6.4, 3.1, 5.5, 1.8],
[6. , 3. , 4.8, 1.8],
[6.9, 3.1, 5.4, 2.1],
[6.7, 3.1, 5.6, 2.4],
[6.9, 3.1, 5.1, 2.3],
[5.8, 2.7, 5.1, 1.9],
[6.8, 3.2, 5.9, 2.3],
[6.7, 3.3, 5.7, 2.5],
[6.7, 3. , 5.2, 2.3],
[6.3, 2.5, 5. , 1.9],
[6.5, 3. , 5.2, 2. ],
[6.2, 3.4, 5.4, 2.3],
[5.9, 3. , 5.1, 1.8]]),
'target': array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2]),
'frame': None,
'target_names': array(['setosa', 'versicolor', 'virginica'], dtype='<U10'),
'DESCR': '.. _iris_dataset:\n\nIris plants dataset\n--------------------\n\n**Data Set Characteristics:**\n\n :Number of Instances: 150 (50 in each of three classes)\n :Number of Attributes: 4 numeric, predictive attributes and the class\n :Attribute Information:\n - sepal length in cm\n - sepal width in cm\n - petal length in cm\n - petal width in cm\n - class:\n - Iris-Setosa\n - Iris-Versicolour\n - Iris-Virginica\n \n :Summary Statistics:\n\n ============== ==== ==== ======= ===== ====================\n Min Max Mean SD Class Correlation\n ============== ==== ==== ======= ===== ====================\n sepal length: 4.3 7.9 5.84 0.83 0.7826\n sepal width: 2.0 4.4 3.05 0.43 -0.4194\n petal length: 1.0 6.9 3.76 1.76 0.9490 (high!)\n petal width: 0.1 2.5 1.20 0.76 0.9565 (high!)\n ============== ==== ==== ======= ===== ====================\n\n :Missing Attribute Values: None\n :Class Distribution: 33.3% for each of 3 classes.\n :Creator: R.A. Fisher\n :Donor: Michael Marshall (MARSHALL%PLU@io.arc.nasa.gov)\n :Date: July, 1988\n\nThe famous Iris database, first used by Sir R.A. Fisher. The dataset is taken\nfrom Fisher\'s paper. Note that it\'s the same as in R, but not as in the UCI\nMachine Learning Repository, which has two wrong data points.\n\nThis is perhaps the best known database to be found in the\npattern recognition literature. Fisher\'s paper is a classic in the field and\nis referenced frequently to this day. (See Duda & Hart, for example.) The\ndata set contains 3 classes of 50 instances each, where each class refers to a\ntype of iris plant. One class is linearly separable from the other 2; the\nlatter are NOT linearly separable from each other.\n\n.. topic:: References\n\n - Fisher, R.A. "The use of multiple measurements in taxonomic problems"\n Annual Eugenics, 7, Part II, 179-188 (1936); also in "Contributions to\n Mathematical Statistics" (John Wiley, NY, 1950).\n - Duda, R.O., & Hart, P.E. (1973) Pattern Classification and Scene Analysis.\n (Q327.D83) John Wiley & Sons. ISBN 0-471-22361-1. See page 218.\n - Dasarathy, B.V. (1980) "Nosing Around the Neighborhood: A New System\n Structure and Classification Rule for Recognition in Partially Exposed\n Environments". IEEE Transactions on Pattern Analysis and Machine\n Intelligence, Vol. PAMI-2, No. 1, 67-71.\n - Gates, G.W. (1972) "The Reduced Nearest Neighbor Rule". IEEE Transactions\n on Information Theory, May 1972, 431-433.\n - See also: 1988 MLC Proceedings, 54-64. Cheeseman et al"s AUTOCLASS II\n conceptual clustering system finds 3 classes in the data.\n - Many, many more ...',
'feature_names': ['sepal length (cm)',
'sepal width (cm)',
'petal length (cm)',
'petal width (cm)'],
'filename': 'iris.csv',
'data_module': 'sklearn.datasets.data'}
print(iris_dataset.DESCR)
.. _iris_dataset:
Iris plants dataset
--------------------
**Data Set Characteristics:**
:Number of Instances: 150 (50 in each of three classes)
:Number of Attributes: 4 numeric, predictive attributes and the class
:Attribute Information:
- sepal length in cm
- sepal width in cm
- petal length in cm
- petal width in cm
- class:
- Iris-Setosa
- Iris-Versicolour
- Iris-Virginica
:Summary Statistics:
============== ==== ==== ======= ===== ====================
Min Max Mean SD Class Correlation
============== ==== ==== ======= ===== ====================
sepal length: 4.3 7.9 5.84 0.83 0.7826
sepal width: 2.0 4.4 3.05 0.43 -0.4194
petal length: 1.0 6.9 3.76 1.76 0.9490 (high!)
petal width: 0.1 2.5 1.20 0.76 0.9565 (high!)
============== ==== ==== ======= ===== ====================
:Missing Attribute Values: None
:Class Distribution: 33.3% for each of 3 classes.
:Creator: R.A. Fisher
:Donor: Michael Marshall (MARSHALL%PLU@io.arc.nasa.gov)
:Date: July, 1988
The famous Iris database, first used by Sir R.A. Fisher. The dataset is taken
from Fisher's paper. Note that it's the same as in R, but not as in the UCI
Machine Learning Repository, which has two wrong data points.
This is perhaps the best known database to be found in the
pattern recognition literature. Fisher's paper is a classic in the field and
is referenced frequently to this day. (See Duda & Hart, for example.) The
data set contains 3 classes of 50 instances each, where each class refers to a
type of iris plant. One class is linearly separable from the other 2; the
latter are NOT linearly separable from each other.
.. topic:: References
- Fisher, R.A. "The use of multiple measurements in taxonomic problems"
Annual Eugenics, 7, Part II, 179-188 (1936); also in "Contributions to
Mathematical Statistics" (John Wiley, NY, 1950).
- Duda, R.O., & Hart, P.E. (1973) Pattern Classification and Scene Analysis.
(Q327.D83) John Wiley & Sons. ISBN 0-471-22361-1. See page 218.
- Dasarathy, B.V. (1980) "Nosing Around the Neighborhood: A New System
Structure and Classification Rule for Recognition in Partially Exposed
Environments". IEEE Transactions on Pattern Analysis and Machine
Intelligence, Vol. PAMI-2, No. 1, 67-71.
- Gates, G.W. (1972) "The Reduced Nearest Neighbor Rule". IEEE Transactions
on Information Theory, May 1972, 431-433.
- See also: 1988 MLC Proceedings, 54-64. Cheeseman et al"s AUTOCLASS II
conceptual clustering system finds 3 classes in the data.
- Many, many more ...
iris_dataset.data
array([[5.1, 3.5, 1.4, 0.2],
[4.9, 3. , 1.4, 0.2],
[4.7, 3.2, 1.3, 0.2],
[4.6, 3.1, 1.5, 0.2],
[5. , 3.6, 1.4, 0.2],
[5.4, 3.9, 1.7, 0.4],
[4.6, 3.4, 1.4, 0.3],
[5. , 3.4, 1.5, 0.2],
[4.4, 2.9, 1.4, 0.2],
[4.9, 3.1, 1.5, 0.1],
[5.4, 3.7, 1.5, 0.2],
[4.8, 3.4, 1.6, 0.2],
[4.8, 3. , 1.4, 0.1],
[4.3, 3. , 1.1, 0.1],
[5.8, 4. , 1.2, 0.2],
[5.7, 4.4, 1.5, 0.4],
[5.4, 3.9, 1.3, 0.4],
[5.1, 3.5, 1.4, 0.3],
[5.7, 3.8, 1.7, 0.3],
[5.1, 3.8, 1.5, 0.3],
[5.4, 3.4, 1.7, 0.2],
[5.1, 3.7, 1.5, 0.4],
[4.6, 3.6, 1. , 0.2],
[5.1, 3.3, 1.7, 0.5],
[4.8, 3.4, 1.9, 0.2],
[5. , 3. , 1.6, 0.2],
[5. , 3.4, 1.6, 0.4],
[5.2, 3.5, 1.5, 0.2],
[5.2, 3.4, 1.4, 0.2],
[4.7, 3.2, 1.6, 0.2],
[4.8, 3.1, 1.6, 0.2],
[5.4, 3.4, 1.5, 0.4],
[5.2, 4.1, 1.5, 0.1],
[5.5, 4.2, 1.4, 0.2],
[4.9, 3.1, 1.5, 0.2],
[5. , 3.2, 1.2, 0.2],
[5.5, 3.5, 1.3, 0.2],
[4.9, 3.6, 1.4, 0.1],
[4.4, 3. , 1.3, 0.2],
[5.1, 3.4, 1.5, 0.2],
[5. , 3.5, 1.3, 0.3],
[4.5, 2.3, 1.3, 0.3],
[4.4, 3.2, 1.3, 0.2],
[5. , 3.5, 1.6, 0.6],
[5.1, 3.8, 1.9, 0.4],
[4.8, 3. , 1.4, 0.3],
[5.1, 3.8, 1.6, 0.2],
[4.6, 3.2, 1.4, 0.2],
[5.3, 3.7, 1.5, 0.2],
[5. , 3.3, 1.4, 0.2],
[7. , 3.2, 4.7, 1.4],
[6.4, 3.2, 4.5, 1.5],
[6.9, 3.1, 4.9, 1.5],
[5.5, 2.3, 4. , 1.3],
[6.5, 2.8, 4.6, 1.5],
[5.7, 2.8, 4.5, 1.3],
[6.3, 3.3, 4.7, 1.6],
[4.9, 2.4, 3.3, 1. ],
[6.6, 2.9, 4.6, 1.3],
[5.2, 2.7, 3.9, 1.4],
[5. , 2. , 3.5, 1. ],
[5.9, 3. , 4.2, 1.5],
[6. , 2.2, 4. , 1. ],
[6.1, 2.9, 4.7, 1.4],
[5.6, 2.9, 3.6, 1.3],
[6.7, 3.1, 4.4, 1.4],
[5.6, 3. , 4.5, 1.5],
[5.8, 2.7, 4.1, 1. ],
[6.2, 2.2, 4.5, 1.5],
[5.6, 2.5, 3.9, 1.1],
[5.9, 3.2, 4.8, 1.8],
[6.1, 2.8, 4. , 1.3],
[6.3, 2.5, 4.9, 1.5],
[6.1, 2.8, 4.7, 1.2],
[6.4, 2.9, 4.3, 1.3],
[6.6, 3. , 4.4, 1.4],
[6.8, 2.8, 4.8, 1.4],
[6.7, 3. , 5. , 1.7],
[6. , 2.9, 4.5, 1.5],
[5.7, 2.6, 3.5, 1. ],
[5.5, 2.4, 3.8, 1.1],
[5.5, 2.4, 3.7, 1. ],
[5.8, 2.7, 3.9, 1.2],
[6. , 2.7, 5.1, 1.6],
[5.4, 3. , 4.5, 1.5],
[6. , 3.4, 4.5, 1.6],
[6.7, 3.1, 4.7, 1.5],
[6.3, 2.3, 4.4, 1.3],
[5.6, 3. , 4.1, 1.3],
[5.5, 2.5, 4. , 1.3],
[5.5, 2.6, 4.4, 1.2],
[6.1, 3. , 4.6, 1.4],
[5.8, 2.6, 4. , 1.2],
[5. , 2.3, 3.3, 1. ],
[5.6, 2.7, 4.2, 1.3],
[5.7, 3. , 4.2, 1.2],
[5.7, 2.9, 4.2, 1.3],
[6.2, 2.9, 4.3, 1.3],
[5.1, 2.5, 3. , 1.1],
[5.7, 2.8, 4.1, 1.3],
[6.3, 3.3, 6. , 2.5],
[5.8, 2.7, 5.1, 1.9],
[7.1, 3. , 5.9, 2.1],
[6.3, 2.9, 5.6, 1.8],
[6.5, 3. , 5.8, 2.2],
[7.6, 3. , 6.6, 2.1],
[4.9, 2.5, 4.5, 1.7],
[7.3, 2.9, 6.3, 1.8],
[6.7, 2.5, 5.8, 1.8],
[7.2, 3.6, 6.1, 2.5],
[6.5, 3.2, 5.1, 2. ],
[6.4, 2.7, 5.3, 1.9],
[6.8, 3. , 5.5, 2.1],
[5.7, 2.5, 5. , 2. ],
[5.8, 2.8, 5.1, 2.4],
[6.4, 3.2, 5.3, 2.3],
[6.5, 3. , 5.5, 1.8],
[7.7, 3.8, 6.7, 2.2],
[7.7, 2.6, 6.9, 2.3],
[6. , 2.2, 5. , 1.5],
[6.9, 3.2, 5.7, 2.3],
[5.6, 2.8, 4.9, 2. ],
[7.7, 2.8, 6.7, 2. ],
[6.3, 2.7, 4.9, 1.8],
[6.7, 3.3, 5.7, 2.1],
[7.2, 3.2, 6. , 1.8],
[6.2, 2.8, 4.8, 1.8],
[6.1, 3. , 4.9, 1.8],
[6.4, 2.8, 5.6, 2.1],
[7.2, 3. , 5.8, 1.6],
[7.4, 2.8, 6.1, 1.9],
[7.9, 3.8, 6.4, 2. ],
[6.4, 2.8, 5.6, 2.2],
[6.3, 2.8, 5.1, 1.5],
[6.1, 2.6, 5.6, 1.4],
[7.7, 3. , 6.1, 2.3],
[6.3, 3.4, 5.6, 2.4],
[6.4, 3.1, 5.5, 1.8],
[6. , 3. , 4.8, 1.8],
[6.9, 3.1, 5.4, 2.1],
[6.7, 3.1, 5.6, 2.4],
[6.9, 3.1, 5.1, 2.3],
[5.8, 2.7, 5.1, 1.9],
[6.8, 3.2, 5.9, 2.3],
[6.7, 3.3, 5.7, 2.5],
[6.7, 3. , 5.2, 2.3],
[6.3, 2.5, 5. , 1.9],
[6.5, 3. , 5.2, 2. ],
[6.2, 3.4, 5.4, 2.3],
[5.9, 3. , 5.1, 1.8]])
iris_dataset.target
array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2])
iris_dataset.target_names
array(['setosa', 'versicolor', 'virginica'], dtype='<U10')
diabetes = load_diabetes()
print(diabetes.DESCR)
.. _diabetes_dataset:
Diabetes dataset
----------------
Ten baseline variables, age, sex, body mass index, average blood
pressure, and six blood serum measurements were obtained for each of n =
442 diabetes patients, as well as the response of interest, a
quantitative measure of disease progression one year after baseline.
**Data Set Characteristics:**
:Number of Instances: 442
:Number of Attributes: First 10 columns are numeric predictive values
:Target: Column 11 is a quantitative measure of disease progression one year after baseline
:Attribute Information:
- age age in years
- sex
- bmi body mass index
- bp average blood pressure
- s1 tc, total serum cholesterol
- s2 ldl, low-density lipoproteins
- s3 hdl, high-density lipoproteins
- s4 tch, total cholesterol / HDL
- s5 ltg, possibly log of serum triglycerides level
- s6 glu, blood sugar level
Note: Each of these 10 feature variables have been mean centered and scaled by the standard deviation times `n_samples` (i.e. the sum of squares of each column totals 1).
Source URL:
https://www4.stat.ncsu.edu/~boos/var.select/diabetes.html
For more information see:
Bradley Efron, Trevor Hastie, Iain Johnstone and Robert Tibshirani (2004) "Least Angle Regression," Annals of Statistics (with discussion), 407-499.
(https://web.stanford.edu/~hastie/Papers/LARS/LeastAngle_2002.pdf)
import pandas as pd
pd.DataFrame(diabetes.data, columns=diabetes.feature_names)
| age | sex | bmi | bp | s1 | s2 | s3 | s4 | s5 | s6 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.038076 | 0.050680 | 0.061696 | 0.021872 | -0.044223 | -0.034821 | -0.043401 | -0.002592 | 0.019908 | -0.017646 |
| 1 | -0.001882 | -0.044642 | -0.051474 | -0.026328 | -0.008449 | -0.019163 | 0.074412 | -0.039493 | -0.068330 | -0.092204 |
| 2 | 0.085299 | 0.050680 | 0.044451 | -0.005671 | -0.045599 | -0.034194 | -0.032356 | -0.002592 | 0.002864 | -0.025930 |
| 3 | -0.089063 | -0.044642 | -0.011595 | -0.036656 | 0.012191 | 0.024991 | -0.036038 | 0.034309 | 0.022692 | -0.009362 |
| 4 | 0.005383 | -0.044642 | -0.036385 | 0.021872 | 0.003935 | 0.015596 | 0.008142 | -0.002592 | -0.031991 | -0.046641 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 437 | 0.041708 | 0.050680 | 0.019662 | 0.059744 | -0.005697 | -0.002566 | -0.028674 | -0.002592 | 0.031193 | 0.007207 |
| 438 | -0.005515 | 0.050680 | -0.015906 | -0.067642 | 0.049341 | 0.079165 | -0.028674 | 0.034309 | -0.018118 | 0.044485 |
| 439 | 0.041708 | 0.050680 | -0.015906 | 0.017282 | -0.037344 | -0.013840 | -0.024993 | -0.011080 | -0.046879 | 0.015491 |
| 440 | -0.045472 | -0.044642 | 0.039062 | 0.001215 | 0.016318 | 0.015283 | -0.028674 | 0.026560 | 0.044528 | -0.025930 |
| 441 | -0.045472 | -0.044642 | -0.073030 | -0.081414 | 0.083740 | 0.027809 | 0.173816 | -0.039493 | -0.004220 | 0.003064 |
442 rows × 10 columns
print(diabetes.DESCR)
.. _diabetes_dataset:
Diabetes dataset
----------------
Ten baseline variables, age, sex, body mass index, average blood
pressure, and six blood serum measurements were obtained for each of n =
442 diabetes patients, as well as the response of interest, a
quantitative measure of disease progression one year after baseline.
**Data Set Characteristics:**
:Number of Instances: 442
:Number of Attributes: First 10 columns are numeric predictive values
:Target: Column 11 is a quantitative measure of disease progression one year after baseline
:Attribute Information:
- age age in years
- sex
- bmi body mass index
- bp average blood pressure
- s1 tc, total serum cholesterol
- s2 ldl, low-density lipoproteins
- s3 hdl, high-density lipoproteins
- s4 tch, total cholesterol / HDL
- s5 ltg, possibly log of serum triglycerides level
- s6 glu, blood sugar level
Note: Each of these 10 feature variables have been mean centered and scaled by the standard deviation times `n_samples` (i.e. the sum of squares of each column totals 1).
Source URL:
https://www4.stat.ncsu.edu/~boos/var.select/diabetes.html
For more information see:
Bradley Efron, Trevor Hastie, Iain Johnstone and Robert Tibshirani (2004) "Least Angle Regression," Annals of Statistics (with discussion), 407-499.
(https://web.stanford.edu/~hastie/Papers/LARS/LeastAngle_2002.pdf)
Loading data from the web#
import sklearn
from sklearn.datasets import fetch_california_housing
houses = fetch_california_housing()
print(houses.DESCR)
.. _california_housing_dataset:
California Housing dataset
--------------------------
**Data Set Characteristics:**
:Number of Instances: 20640
:Number of Attributes: 8 numeric, predictive attributes and the target
:Attribute Information:
- MedInc median income in block group
- HouseAge median house age in block group
- AveRooms average number of rooms per household
- AveBedrms average number of bedrooms per household
- Population block group population
- AveOccup average number of household members
- Latitude block group latitude
- Longitude block group longitude
:Missing Attribute Values: None
This dataset was obtained from the StatLib repository.
https://www.dcc.fc.up.pt/~ltorgo/Regression/cal_housing.html
The target variable is the median house value for California districts,
expressed in hundreds of thousands of dollars ($100,000).
This dataset was derived from the 1990 U.S. census, using one row per census
block group. A block group is the smallest geographical unit for which the U.S.
Census Bureau publishes sample data (a block group typically has a population
of 600 to 3,000 people).
An household is a group of people residing within a home. Since the average
number of rooms and bedrooms in this dataset are provided per household, these
columns may take surpinsingly large values for block groups with few households
and many empty houses, such as vacation resorts.
It can be downloaded/loaded using the
:func:`sklearn.datasets.fetch_california_housing` function.
.. topic:: References
- Pace, R. Kelley and Ronald Barry, Sparse Spatial Autoregressions,
Statistics and Probability Letters, 33 (1997) 291-297
houses.data.shape
houses.feature_names
pd.DataFrame(houses.data, columns=houses.feature_names)
(20640, 8)
['MedInc',
'HouseAge',
'AveRooms',
'AveBedrms',
'Population',
'AveOccup',
'Latitude',
'Longitude']
| MedInc | HouseAge | AveRooms | AveBedrms | Population | AveOccup | Latitude | Longitude | |
|---|---|---|---|---|---|---|---|---|
| 0 | 8.3252 | 41.0 | 6.984127 | 1.023810 | 322.0 | 2.555556 | 37.88 | -122.23 |
| 1 | 8.3014 | 21.0 | 6.238137 | 0.971880 | 2401.0 | 2.109842 | 37.86 | -122.22 |
| 2 | 7.2574 | 52.0 | 8.288136 | 1.073446 | 496.0 | 2.802260 | 37.85 | -122.24 |
| 3 | 5.6431 | 52.0 | 5.817352 | 1.073059 | 558.0 | 2.547945 | 37.85 | -122.25 |
| 4 | 3.8462 | 52.0 | 6.281853 | 1.081081 | 565.0 | 2.181467 | 37.85 | -122.25 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 20635 | 1.5603 | 25.0 | 5.045455 | 1.133333 | 845.0 | 2.560606 | 39.48 | -121.09 |
| 20636 | 2.5568 | 18.0 | 6.114035 | 1.315789 | 356.0 | 3.122807 | 39.49 | -121.21 |
| 20637 | 1.7000 | 17.0 | 5.205543 | 1.120092 | 1007.0 | 2.325635 | 39.43 | -121.22 |
| 20638 | 1.8672 | 18.0 | 5.329513 | 1.171920 | 741.0 | 2.123209 | 39.43 | -121.32 |
| 20639 | 2.3886 | 16.0 | 5.254717 | 1.162264 | 1387.0 | 2.616981 | 39.37 | -121.24 |
20640 rows × 8 columns
diabetes.data.shape
(442, 10)
Generate dataset#
from sklearn.datasets import make_regression
x,y = make_regression(n_samples=100, n_features=5, n_targets=1, noise=0.005)
pd.DataFrame(y)
| 0 | |
|---|---|
| 0 | -56.878301 |
| 1 | 157.174068 |
| 2 | -20.096663 |
| 3 | -7.563574 |
| 4 | 27.367709 |
| ... | ... |
| 95 | -31.741801 |
| 96 | 8.639267 |
| 97 | -68.500575 |
| 98 | -62.879911 |
| 99 | -10.838747 |
100 rows × 1 columns
pd.DataFrame(x)
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| 0 | 0.161720 | 0.091843 | 0.095730 | -1.913515 | 0.140882 |
| 1 | 0.272338 | 1.283241 | 0.529606 | 0.973450 | 0.628376 |
| 2 | -0.251843 | -0.157911 | 1.327633 | -1.302578 | 0.441988 |
| 3 | 0.117548 | 0.301254 | -0.043708 | 0.134207 | -0.478126 |
| 4 | -1.040602 | 0.352708 | 0.360921 | -1.766644 | 1.595943 |
| ... | ... | ... | ... | ... | ... |
| 95 | 0.558344 | -0.631148 | -1.208268 | -0.654117 | 0.435893 |
| 96 | 0.971506 | -0.573440 | 0.403751 | 0.455868 | -0.173494 |
| 97 | -0.506183 | -0.375183 | 1.588506 | -0.896150 | -0.352137 |
| 98 | 0.186767 | 0.093694 | -1.776359 | 0.301386 | -0.949856 |
| 99 | -0.102822 | -0.613607 | -1.191659 | 2.246375 | -0.792975 |
100 rows × 5 columns
import seaborn as sns
sns.set(color_codes=True)
sns.regplot(x=x, y=y);
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Input In [21], in <cell line: 3>()
1 import seaborn as sns
2 sns.set(color_codes=True)
----> 3 sns.regplot(x=x, y=y)
File ~/anaconda3/lib/python3.9/site-packages/seaborn/_decorators.py:46, in _deprecate_positional_args.<locals>.inner_f(*args, **kwargs)
36 warnings.warn(
37 "Pass the following variable{} as {}keyword arg{}: {}. "
38 "From version 0.12, the only valid positional argument "
(...)
43 FutureWarning
44 )
45 kwargs.update({k: arg for k, arg in zip(sig.parameters, args)})
---> 46 return f(**kwargs)
File ~/anaconda3/lib/python3.9/site-packages/seaborn/regression.py:851, in regplot(x, y, data, x_estimator, x_bins, x_ci, scatter, fit_reg, ci, n_boot, units, seed, order, logistic, lowess, robust, logx, x_partial, y_partial, truncate, dropna, x_jitter, y_jitter, label, color, marker, scatter_kws, line_kws, ax)
837 @_deprecate_positional_args
838 def regplot(
839 *,
(...)
848 scatter_kws=None, line_kws=None, ax=None
849 ):
--> 851 plotter = _RegressionPlotter(x, y, data, x_estimator, x_bins, x_ci,
852 scatter, fit_reg, ci, n_boot, units, seed,
853 order, logistic, lowess, robust, logx,
854 x_partial, y_partial, truncate, dropna,
855 x_jitter, y_jitter, color, label)
857 if ax is None:
858 ax = plt.gca()
File ~/anaconda3/lib/python3.9/site-packages/seaborn/regression.py:109, in _RegressionPlotter.__init__(self, x, y, data, x_estimator, x_bins, x_ci, scatter, fit_reg, ci, n_boot, units, seed, order, logistic, lowess, robust, logx, x_partial, y_partial, truncate, dropna, x_jitter, y_jitter, color, label)
106 raise ValueError("Mutually exclusive regression options.")
108 # Extract the data vals from the arguments or passed dataframe
--> 109 self.establish_variables(data, x=x, y=y, units=units,
110 x_partial=x_partial, y_partial=y_partial)
112 # Drop null observations
113 if dropna:
File ~/anaconda3/lib/python3.9/site-packages/seaborn/regression.py:55, in _LinearPlotter.establish_variables(self, data, **kws)
53 if np.ndim(vector) > 1:
54 err = "regplot inputs must be 1d"
---> 55 raise ValueError(err)
56 setattr(self, var, vector)
ValueError: regplot inputs must be 1d
Load data from openml.org#
from sklearn.datasets import fetch_openml
mice = fetch_openml(name='miceprotein')
print(mice.target)
0 c-CS-m
1 c-CS-m
2 c-CS-m
3 c-CS-m
4 c-CS-m
...
1075 t-SC-s
1076 t-SC-s
1077 t-SC-s
1078 t-SC-s
1079 t-SC-s
Name: class, Length: 1080, dtype: category
Categories (8, object): ['c-CS-m', 'c-CS-s', 'c-SC-m', 'c-SC-s', 't-CS-m', 't-CS-s', 't-SC-m', 't-SC-s']
import numpy as np
d = np.random.rand(3,10)
d.shape
mice.data.shape
print(mice.DESCR)
mice.target_names
import pandas as pd
5.3. Preprocessing data#
Standardization, or mean removal and variance scaling#
Standardization of datasets is a common requirement for many machine learning estimators implemented in scikit-learn; they might behave badly if the individual features do not more or less look like standard normally distributed data: Gaussian with zero mean and unit variance.
In practice we often ignore the shape of the distribution and just transform the data to center it by removing the mean value of each feature, then scale it by dividing non-constant features by their standard deviation.
from IPython.display import display
from sklearn import preprocessing
import numpy as np
X_train = np.array([[ 1., -1., 2.],
[ 2., 0., 0.],
[ 0., 1., -1.]])
df = pd.DataFrame(X_train)
display(df)
X_scaled = preprocessing.scale(df)
X_scaled
#rows are different patients/subjects/flowers
# columns are various features of the patients/subjects/flowers
| 0 | 1 | 2 | |
|---|---|---|---|
| 0 | 1.0 | -1.0 | 2.0 |
| 1 | 2.0 | 0.0 | 0.0 |
| 2 | 0.0 | 1.0 | -1.0 |
array([[ 0. , -1.22474487, 1.33630621],
[ 1.22474487, 0. , -0.26726124],
[-1.22474487, 1.22474487, -1.06904497]])
df.describe()
| 0 | 1 | 2 | |
|---|---|---|---|
| count | 3.0 | 3.0 | 3.000000 |
| mean | 1.0 | 0.0 | 0.333333 |
| std | 1.0 | 1.0 | 1.527525 |
| min | 0.0 | -1.0 | -1.000000 |
| 25% | 0.5 | -0.5 | -0.500000 |
| 50% | 1.0 | 0.0 | 0.000000 |
| 75% | 1.5 | 0.5 | 1.000000 |
| max | 2.0 | 1.0 | 2.000000 |
sum([0. , -1.22474487, 1.33630621])
0.11156133999999995
idata = iris_dataset.data
id_scaled = preprocessing.scale(idata)
idata.mean(axis=0)
id_scaled.mean(axis=0)
id_scaled.var(axis=0)
array([5.84333333, 3.05733333, 3.758 , 1.19933333])
array([-1.69031455e-15, -1.84297022e-15, -1.69864123e-15, -1.40924309e-15])
array([1., 1., 1., 1.])
X_train.mean(axis=0)
X_scaled.mean(axis=0)
array([1. , 0. , 0.33333333])
array([0., 0., 0.])
X_scaled.std(axis=0)
X_scaled.mean(axis=1)
X_scaled.std(axis=1)
Can save scaling and apply to testing data#
scaler = preprocessing.StandardScaler().fit(X_train)
scaler
scaler.transform(X_train)
StandardScaler()
array([[ 0. , -1.22474487, 1.33630621],
[ 1.22474487, 0. , -0.26726124],
[-1.22474487, 1.22474487, -1.06904497]])
scaler.get_params
{'copy': True, 'with_mean': True, 'with_std': True}
X_test = [[-1., 1., 0.]]
scaler.transform(X_test)
array([[-2.44948974, 1.22474487, -0.26726124]])
Scaling features to a range#
X_train = np.array([[ 1., -1., 2.],
[ 2., 0., 0.],
[ 0., 1., -1.]])
min_max_scaler = preprocessing.MinMaxScaler()
X_train_minmax = min_max_scaler.fit_transform(X_train)
X_train_minmax
array([[0.5 , 0. , 1. ],
[1. , 0.5 , 0.33333333],
[0. , 1. , 0. ]])
X_test = np.array([[-3., -1., 4.]])
X_test_minmax = min_max_scaler.transform(X_test)
X_test_minmax
array([[-1.5 , 0. , 1.66666667]])
Pre-processing data - Non-linear transformation#
import pandas as pd
%matplotlib inline
df = pd.read_csv('international-airline-passengers.csv')
display(df)
| date | passengers | |
|---|---|---|
| 0 | 1949-01 | 112.0 |
| 1 | 1949-02 | 118.0 |
| 2 | 1949-03 | 132.0 |
| 3 | 1949-04 | 129.0 |
| 4 | 1949-05 | 121.0 |
| ... | ... | ... |
| 140 | 1960-09 | 508.0 |
| 141 | 1960-10 | 461.0 |
| 142 | 1960-11 | 390.0 |
| 143 | 1960-12 | 432.0 |
| 144 | International airline passengers: monthly tota... | NaN |
145 rows × 2 columns
df.keys()
Index(['date', 'passengers'], dtype='object')
df['passengers'].hist(bins=20)
<AxesSubplot:>
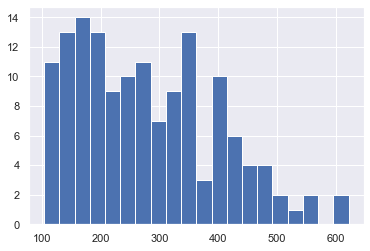
import numpy as np
df['passengers'] = np.log(df['passengers'])
df['passengers'].hist(bins=20)
<AxesSubplot:>
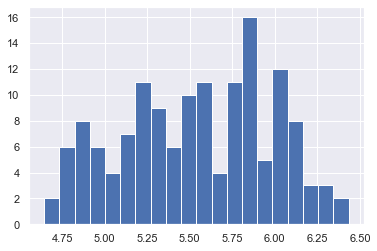
5.3.3. Normalization#
from sklearn import preprocessing
X = [[ 1., -1., 2.],
[ 2., 0., 0.],
[ 0., 1., -1.]]
X_normalized = preprocessing.normalize(X, norm='l1')
X_normalized
# http://www.chioka.in/differences-between-the-l1-norm-and-the-l2-norm-least-absolute-deviations-and-least-squares/
array([[ 0.25, -0.25, 0.5 ],
[ 1. , 0. , 0. ],
[ 0. , 0.5 , -0.5 ]])
Can save the normalization for future use#
normalizer = preprocessing.Normalizer().fit(X) # fit does nothing
normalizer
Normalizer()
normalizer.transform(X)
array([[ 0.40824829, -0.40824829, 0.81649658],
[ 1. , 0. , 0. ],
[ 0. , 0.70710678, -0.70710678]])
tmp = normalizer.transform([[2., 1., 0.]])
tmp
array([[0.89442719, 0.4472136 , 0. ]])
tmp.mean()
0.4472135954999579
(tmp*tmp).sum()
0.9999999999999999
Preprocessing data - Encoding#
from sklearn import preprocessing
enc = preprocessing.OrdinalEncoder()
X = [['male', 'from US', 'uses Safari'],
['female', 'from Europe', 'uses Firefox']]
enc.fit(X)
OrdinalEncoder()
enc.transform([['female', 'from US', 'uses Safari']])
array([[0., 1., 1.]])
enc.transform([['male', 'from Europe', 'uses Safari']])
array([[1., 0., 1.]])
enc.transform([['female', 'from Europe', 'uses Firefox']])
array([[0., 0., 0.]])
genders = ['female', 'male']
locations = ['from Africa', 'from Asia', 'from Europe', 'from US']
browsers = ['uses Chrome', 'uses Firefox', 'uses IE', 'uses Safari']
enc = preprocessing.OneHotEncoder(categories=[genders, locations, browsers])
X = [['male', 'from US', 'uses Safari'], ['female', 'from Europe', 'uses Firefox']]
enc.fit(X)
enc.categories_
OneHotEncoder(categories=[['female', 'male'],
['from Africa', 'from Asia', 'from Europe',
'from US'],
['uses Chrome', 'uses Firefox', 'uses IE',
'uses Safari']])
[array(['female', 'male'], dtype=object),
array(['from Africa', 'from Asia', 'from Europe', 'from US'], dtype=object),
array(['uses Chrome', 'uses Firefox', 'uses IE', 'uses Safari'],
dtype=object)]
enc.transform([['male', 'from US', 'uses Safari']])
tmp = enc.transform([['female', 'from Asia', 'uses Chrome'],
['male', 'from Europe', 'uses Safari']]).toarray()
tmp
array([[1., 0., 0., 1., 0., 0., 1., 0., 0., 0.],
[0., 1., 0., 0., 1., 0., 0., 0., 0., 1.]])
[0, 1, 0, 0, 1, 0, 0, 0, 0, 1]
[0, 1, 0, 0, 1, 0, 0, 0, 0, 1]
enc.inverse_transform(tmp)
array([['female', 'from Asia', 'uses Chrome'],
['male', 'from Europe', 'uses Safari']], dtype=object)
5.3.5. Discretization#
Discretization (otherwise known as quantization or binning) provides a way to partition continuous features into discrete values. Certain datasets with continuous features may benefit from discretization, because discretization can transform the dataset of continuous attributes to one with only nominal attributes.
One-hot encoded discretized features can make a model more expressive, while maintaining interpretability. For instance, pre-processing with a discretizer can introduce nonlinearity to linear models.
X = np.array([[ -3., 5., 15 ],
[ 0., 6., 14 ],
[ 6., 3., 11 ]])
est = preprocessing.KBinsDiscretizer(n_bins=[3, 2, 4], encode='ordinal').fit(X)
est.transform(X)
array([[0., 1., 3.],
[1., 1., 2.],
[2., 0., 0.]])
#https://scikit-learn.org/stable/modules/preprocessing.html#k-bins-discretization
5.4.2. Univariate feature imputation#
# Example 1
import numpy as np
from sklearn.impute import SimpleImputer
imp = SimpleImputer(missing_values=np.nan, strategy='mean')
orig_data = [[1, 2],
[np.nan, 3],
[7, 6]]
imp.fit(orig_data)
imp.transform(orig_data)
SimpleImputer()
array([[1., 2.],
[4., 3.],
[7., 6.]])
X = [[np.nan, 2],
[6, np.nan],
[7, 6]]
print(imp.transform(X))
[[4. 2. ]
[6. 3.66666667]
[7. 6. ]]
11/3
3.6666666666666665
import pandas as pd
df = pd.DataFrame([["a", "x"],
[np.nan, "w"],
["a", np.nan],
["b", "y"]], dtype="category")
imp = SimpleImputer(strategy="most_frequent")
print(imp.fit_transform(df))
[['a' 'x']
['a' 'w']
['a' 'w']
['b' 'y']]
import pandas as pd
df = pd.DataFrame([["a", "x"],
[np.nan, "y"],
["c", np.nan],
["b", "y"]], dtype="category")
imp = SimpleImputer(strategy="most_frequent")
print(imp.fit_transform(df))
[['a' 'x']
['a' 'y']
['c' 'y']
['b' 'y']]
Splitting data into Train and Test#
import numpy as np
from sklearn.model_selection import train_test_split
X, y = np.arange(10).reshape((5, 2)), [0, 1, 0, 0, 1]
X
list(y)
# X -- feature
# y -- label
array([[0, 1],
[2, 3],
[4, 5],
[6, 7],
[8, 9]])
[0, 1, 0, 0, 1]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
X_train
y_train
X_test
y_test
array([[4, 5],
[2, 3],
[0, 1],
[8, 9]])
[0, 1, 0, 1]
array([[6, 7]])
[0]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.40, random_state=43)
X_train
y_train
X_test
y_test
array([[2, 3],
[0, 1],
[8, 9]])
[1, 0, 1]
array([[6, 7],
[4, 5]])
[0, 0]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=42)
X_train
y_train
X_test
y_test
array([[4, 5],
[0, 1],
[6, 7]])
[0, 0, 0]
array([[2, 3],
[8, 9]])
[1, 1]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=np.random)
X_train
y_train
X_test
y_test
array([[6, 7],
[8, 9],
[4, 5]])
[0, 1, 0]
array([[0, 1],
[2, 3]])
[0, 1]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=np.random)
X_train
y_train
X_test
y_test
array([[0, 1],
[6, 7],
[8, 9]])
[0, 0, 1]
array([[2, 3],
[4, 5]])
[1, 0]
import pandas as pd
from sklearn import datasets, linear_model
from sklearn.model_selection import train_test_split
from matplotlib import pyplot as plt
diabetes = datasets.load_diabetes()
diabetes.data.shape
(442, 10)
feature_names = diabetes.feature_names
feature_names
['age', 'sex', 'bmi', 'bp', 's1', 's2', 's3', 's4', 's5', 's6']
print(diabetes.DESCR)
df = pd.DataFrame(diabetes.data, columns=feature_names)
y = diabetes.target
df
y
| age | sex | bmi | bp | s1 | s2 | s3 | s4 | s5 | s6 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.038076 | 0.050680 | 0.061696 | 0.021872 | -0.044223 | -0.034821 | -0.043401 | -0.002592 | 0.019908 | -0.017646 |
| 1 | -0.001882 | -0.044642 | -0.051474 | -0.026328 | -0.008449 | -0.019163 | 0.074412 | -0.039493 | -0.068330 | -0.092204 |
| 2 | 0.085299 | 0.050680 | 0.044451 | -0.005671 | -0.045599 | -0.034194 | -0.032356 | -0.002592 | 0.002864 | -0.025930 |
| 3 | -0.089063 | -0.044642 | -0.011595 | -0.036656 | 0.012191 | 0.024991 | -0.036038 | 0.034309 | 0.022692 | -0.009362 |
| 4 | 0.005383 | -0.044642 | -0.036385 | 0.021872 | 0.003935 | 0.015596 | 0.008142 | -0.002592 | -0.031991 | -0.046641 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 437 | 0.041708 | 0.050680 | 0.019662 | 0.059744 | -0.005697 | -0.002566 | -0.028674 | -0.002592 | 0.031193 | 0.007207 |
| 438 | -0.005515 | 0.050680 | -0.015906 | -0.067642 | 0.049341 | 0.079165 | -0.028674 | 0.034309 | -0.018118 | 0.044485 |
| 439 | 0.041708 | 0.050680 | -0.015906 | 0.017282 | -0.037344 | -0.013840 | -0.024993 | -0.011080 | -0.046879 | 0.015491 |
| 440 | -0.045472 | -0.044642 | 0.039062 | 0.001215 | 0.016318 | 0.015283 | -0.028674 | 0.026560 | 0.044528 | -0.025930 |
| 441 | -0.045472 | -0.044642 | -0.073030 | -0.081414 | 0.083740 | 0.027809 | 0.173816 | -0.039493 | -0.004220 | 0.003064 |
442 rows × 10 columns
array([151., 75., 141., 206., 135., 97., 138., 63., 110., 310., 101.,
69., 179., 185., 118., 171., 166., 144., 97., 168., 68., 49.,
68., 245., 184., 202., 137., 85., 131., 283., 129., 59., 341.,
87., 65., 102., 265., 276., 252., 90., 100., 55., 61., 92.,
259., 53., 190., 142., 75., 142., 155., 225., 59., 104., 182.,
128., 52., 37., 170., 170., 61., 144., 52., 128., 71., 163.,
150., 97., 160., 178., 48., 270., 202., 111., 85., 42., 170.,
200., 252., 113., 143., 51., 52., 210., 65., 141., 55., 134.,
42., 111., 98., 164., 48., 96., 90., 162., 150., 279., 92.,
83., 128., 102., 302., 198., 95., 53., 134., 144., 232., 81.,
104., 59., 246., 297., 258., 229., 275., 281., 179., 200., 200.,
173., 180., 84., 121., 161., 99., 109., 115., 268., 274., 158.,
107., 83., 103., 272., 85., 280., 336., 281., 118., 317., 235.,
60., 174., 259., 178., 128., 96., 126., 288., 88., 292., 71.,
197., 186., 25., 84., 96., 195., 53., 217., 172., 131., 214.,
59., 70., 220., 268., 152., 47., 74., 295., 101., 151., 127.,
237., 225., 81., 151., 107., 64., 138., 185., 265., 101., 137.,
143., 141., 79., 292., 178., 91., 116., 86., 122., 72., 129.,
142., 90., 158., 39., 196., 222., 277., 99., 196., 202., 155.,
77., 191., 70., 73., 49., 65., 263., 248., 296., 214., 185.,
78., 93., 252., 150., 77., 208., 77., 108., 160., 53., 220.,
154., 259., 90., 246., 124., 67., 72., 257., 262., 275., 177.,
71., 47., 187., 125., 78., 51., 258., 215., 303., 243., 91.,
150., 310., 153., 346., 63., 89., 50., 39., 103., 308., 116.,
145., 74., 45., 115., 264., 87., 202., 127., 182., 241., 66.,
94., 283., 64., 102., 200., 265., 94., 230., 181., 156., 233.,
60., 219., 80., 68., 332., 248., 84., 200., 55., 85., 89.,
31., 129., 83., 275., 65., 198., 236., 253., 124., 44., 172.,
114., 142., 109., 180., 144., 163., 147., 97., 220., 190., 109.,
191., 122., 230., 242., 248., 249., 192., 131., 237., 78., 135.,
244., 199., 270., 164., 72., 96., 306., 91., 214., 95., 216.,
263., 178., 113., 200., 139., 139., 88., 148., 88., 243., 71.,
77., 109., 272., 60., 54., 221., 90., 311., 281., 182., 321.,
58., 262., 206., 233., 242., 123., 167., 63., 197., 71., 168.,
140., 217., 121., 235., 245., 40., 52., 104., 132., 88., 69.,
219., 72., 201., 110., 51., 277., 63., 118., 69., 273., 258.,
43., 198., 242., 232., 175., 93., 168., 275., 293., 281., 72.,
140., 189., 181., 209., 136., 261., 113., 131., 174., 257., 55.,
84., 42., 146., 212., 233., 91., 111., 152., 120., 67., 310.,
94., 183., 66., 173., 72., 49., 64., 48., 178., 104., 132.,
220., 57.])
diabetes.target
array([151., 75., 141., 206., 135., 97., 138., 63., 110., 310., 101.,
69., 179., 185., 118., 171., 166., 144., 97., 168., 68., 49.,
68., 245., 184., 202., 137., 85., 131., 283., 129., 59., 341.,
87., 65., 102., 265., 276., 252., 90., 100., 55., 61., 92.,
259., 53., 190., 142., 75., 142., 155., 225., 59., 104., 182.,
128., 52., 37., 170., 170., 61., 144., 52., 128., 71., 163.,
150., 97., 160., 178., 48., 270., 202., 111., 85., 42., 170.,
200., 252., 113., 143., 51., 52., 210., 65., 141., 55., 134.,
42., 111., 98., 164., 48., 96., 90., 162., 150., 279., 92.,
83., 128., 102., 302., 198., 95., 53., 134., 144., 232., 81.,
104., 59., 246., 297., 258., 229., 275., 281., 179., 200., 200.,
173., 180., 84., 121., 161., 99., 109., 115., 268., 274., 158.,
107., 83., 103., 272., 85., 280., 336., 281., 118., 317., 235.,
60., 174., 259., 178., 128., 96., 126., 288., 88., 292., 71.,
197., 186., 25., 84., 96., 195., 53., 217., 172., 131., 214.,
59., 70., 220., 268., 152., 47., 74., 295., 101., 151., 127.,
237., 225., 81., 151., 107., 64., 138., 185., 265., 101., 137.,
143., 141., 79., 292., 178., 91., 116., 86., 122., 72., 129.,
142., 90., 158., 39., 196., 222., 277., 99., 196., 202., 155.,
77., 191., 70., 73., 49., 65., 263., 248., 296., 214., 185.,
78., 93., 252., 150., 77., 208., 77., 108., 160., 53., 220.,
154., 259., 90., 246., 124., 67., 72., 257., 262., 275., 177.,
71., 47., 187., 125., 78., 51., 258., 215., 303., 243., 91.,
150., 310., 153., 346., 63., 89., 50., 39., 103., 308., 116.,
145., 74., 45., 115., 264., 87., 202., 127., 182., 241., 66.,
94., 283., 64., 102., 200., 265., 94., 230., 181., 156., 233.,
60., 219., 80., 68., 332., 248., 84., 200., 55., 85., 89.,
31., 129., 83., 275., 65., 198., 236., 253., 124., 44., 172.,
114., 142., 109., 180., 144., 163., 147., 97., 220., 190., 109.,
191., 122., 230., 242., 248., 249., 192., 131., 237., 78., 135.,
244., 199., 270., 164., 72., 96., 306., 91., 214., 95., 216.,
263., 178., 113., 200., 139., 139., 88., 148., 88., 243., 71.,
77., 109., 272., 60., 54., 221., 90., 311., 281., 182., 321.,
58., 262., 206., 233., 242., 123., 167., 63., 197., 71., 168.,
140., 217., 121., 235., 245., 40., 52., 104., 132., 88., 69.,
219., 72., 201., 110., 51., 277., 63., 118., 69., 273., 258.,
43., 198., 242., 232., 175., 93., 168., 275., 293., 281., 72.,
140., 189., 181., 209., 136., 261., 113., 131., 174., 257., 55.,
84., 42., 146., 212., 233., 91., 111., 152., 120., 67., 310.,
94., 183., 66., 173., 72., 49., 64., 48., 178., 104., 132.,
220., 57.])
df.shape
(442, 10)
len(y)
X_train, X_test, y_train, y_test = train_test_split(df, y, test_size=0.2, random_state=np.random)
X_train
y_train
X_test
y_test
| age | sex | bmi | bp | s1 | s2 | s3 | s4 | s5 | s6 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 234 | 0.045341 | -0.044642 | 0.039062 | 0.045972 | 0.006687 | -0.024174 | 0.008142 | -0.012556 | 0.064328 | 0.056912 |
| 284 | 0.041708 | 0.050680 | -0.022373 | 0.028758 | -0.066239 | -0.045155 | -0.061809 | -0.002592 | 0.002864 | -0.054925 |
| 242 | -0.103593 | 0.050680 | -0.023451 | -0.022885 | -0.086878 | -0.067701 | -0.017629 | -0.039493 | -0.078141 | -0.071494 |
| 188 | 0.005383 | -0.044642 | -0.002973 | 0.049415 | 0.074108 | 0.070710 | 0.044958 | -0.002592 | -0.001499 | -0.009362 |
| 394 | 0.034443 | -0.044642 | 0.018584 | 0.056301 | 0.012191 | -0.054549 | -0.069172 | 0.071210 | 0.130081 | 0.007207 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 412 | 0.074401 | -0.044642 | 0.085408 | 0.063187 | 0.014942 | 0.013091 | 0.015505 | -0.002592 | 0.006209 | 0.085907 |
| 3 | -0.089063 | -0.044642 | -0.011595 | -0.036656 | 0.012191 | 0.024991 | -0.036038 | 0.034309 | 0.022692 | -0.009362 |
| 83 | -0.038207 | -0.044642 | 0.009961 | -0.046985 | -0.059359 | -0.052983 | -0.010266 | -0.039493 | -0.015998 | -0.042499 |
| 301 | -0.001882 | 0.050680 | -0.024529 | 0.052858 | 0.027326 | 0.030001 | 0.030232 | -0.002592 | -0.021394 | 0.036201 |
| 359 | 0.038076 | 0.050680 | 0.005650 | 0.032201 | 0.006687 | 0.017475 | -0.024993 | 0.034309 | 0.014823 | 0.061054 |
353 rows × 10 columns
array([246., 156., 71., 141., 273., 63., 174., 244., 70., 201., 31.,
108., 303., 77., 89., 158., 181., 118., 53., 220., 54., 72.,
109., 217., 78., 53., 150., 225., 88., 142., 45., 115., 48.,
137., 91., 302., 111., 49., 103., 160., 80., 173., 140., 84.,
161., 69., 132., 88., 59., 249., 190., 74., 295., 128., 122.,
237., 235., 71., 63., 87., 59., 220., 64., 179., 178., 248.,
186., 191., 122., 93., 90., 63., 171., 96., 77., 104., 248.,
72., 341., 230., 144., 93., 235., 202., 292., 151., 120., 58.,
178., 92., 263., 252., 196., 42., 39., 236., 277., 129., 100.,
77., 200., 40., 166., 220., 63., 178., 61., 60., 306., 154.,
47., 147., 94., 96., 202., 216., 258., 275., 257., 42., 222.,
50., 262., 48., 109., 253., 283., 65., 264., 200., 150., 110.,
160., 265., 180., 233., 64., 37., 67., 136., 72., 245., 85.,
102., 172., 69., 99., 61., 73., 128., 172., 246., 81., 111.,
137., 48., 52., 196., 215., 242., 65., 109., 185., 131., 69.,
174., 265., 75., 310., 346., 98., 139., 121., 168., 258., 190.,
118., 132., 79., 178., 292., 52., 111., 242., 150., 129., 297.,
212., 104., 95., 143., 84., 49., 155., 310., 214., 143., 77.,
146., 97., 272., 184., 144., 47., 138., 49., 279., 252., 134.,
91., 89., 200., 153., 72., 163., 170., 51., 276., 170., 142.,
168., 281., 197., 198., 232., 53., 52., 94., 288., 121., 259.,
141., 152., 138., 72., 275., 151., 281., 43., 126., 81., 101.,
59., 110., 127., 94., 134., 139., 155., 281., 206., 131., 200.,
262., 277., 114., 74., 237., 66., 198., 71., 71., 252., 55.,
107., 68., 101., 242., 104., 177., 180., 124., 125., 192., 308.,
51., 116., 68., 263., 163., 243., 181., 150., 60., 91., 59.,
65., 198., 101., 113., 84., 96., 70., 170., 64., 214., 195.,
140., 90., 321., 115., 152., 71., 131., 66., 191., 67., 25.,
72., 199., 127., 135., 336., 175., 42., 219., 245., 221., 75.,
124., 187., 248., 128., 162., 44., 107., 233., 281., 57., 95.,
178., 131., 113., 200., 168., 183., 84., 85., 68., 257., 123.,
53., 116., 283., 83., 275., 151., 118., 261., 206., 210., 65.,
311.])
| age | sex | bmi | bp | s1 | s2 | s3 | s4 | s5 | s6 | |
|---|---|---|---|---|---|---|---|---|---|---|
| 438 | -0.005515 | 0.050680 | -0.015906 | -0.067642 | 0.049341 | 0.079165 | -0.028674 | 0.034309 | -0.018118 | 0.044485 |
| 410 | -0.009147 | 0.050680 | -0.027762 | 0.008101 | 0.047965 | 0.037203 | -0.028674 | 0.034309 | 0.066048 | -0.042499 |
| 44 | 0.045341 | 0.050680 | 0.068163 | 0.008101 | -0.016704 | 0.004636 | -0.076536 | 0.071210 | 0.032433 | -0.017646 |
| 220 | 0.023546 | 0.050680 | -0.039618 | -0.005671 | -0.048351 | -0.033255 | 0.011824 | -0.039493 | -0.101644 | -0.067351 |
| 18 | -0.038207 | -0.044642 | -0.010517 | -0.036656 | -0.037344 | -0.019476 | -0.028674 | -0.002592 | -0.018118 | -0.017646 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 375 | 0.045341 | 0.050680 | -0.002973 | 0.107944 | 0.035582 | 0.022485 | 0.026550 | -0.002592 | 0.028017 | 0.019633 |
| 295 | -0.052738 | 0.050680 | 0.039062 | -0.040099 | -0.005697 | -0.012900 | 0.011824 | -0.039493 | 0.016305 | 0.003064 |
| 126 | -0.089063 | -0.044642 | -0.061174 | -0.026328 | -0.055231 | -0.054549 | 0.041277 | -0.076395 | -0.093936 | -0.054925 |
| 168 | 0.001751 | 0.050680 | 0.059541 | -0.002228 | 0.061725 | 0.063195 | -0.058127 | 0.108111 | 0.068982 | 0.127328 |
| 63 | -0.034575 | -0.044642 | -0.037463 | -0.060757 | 0.020446 | 0.043466 | -0.013948 | -0.002592 | -0.030751 | -0.071494 |
89 rows × 10 columns
array([104., 209., 259., 78., 97., 164., 96., 113., 272., 102., 83.,
51., 60., 90., 167., 55., 259., 229., 90., 145., 310., 265.,
189., 270., 317., 214., 220., 243., 103., 97., 241., 185., 142.,
148., 55., 258., 225., 164., 78., 52., 296., 173., 102., 280.,
232., 182., 87., 230., 208., 109., 129., 200., 233., 92., 83.,
274., 144., 144., 219., 88., 179., 91., 85., 182., 97., 202.,
293., 202., 197., 275., 142., 55., 158., 86., 332., 182., 88.,
39., 270., 185., 268., 135., 90., 141., 217., 85., 99., 268.,
128.])
X_train.shape
len(y_train)
X_test.shape
len(y_test)
(353, 10)
353
(89, 10)
89
Linear Regression#
Ref: https://www.kaggle.com/getting-started/59856
import numpy as np
from sklearn.linear_model import LinearRegression
X = np.array([[1, 1], [1, 2], [2, 2], [2, 3]])
# y = 1 * x_0 + 2 * x_1 + 3
y = np.dot(X, np.array([1, 2])) + 3
reg = LinearRegression().fit(X, y)
reg.score(X, y)
reg.coef_
reg.intercept_
# y = mx + b
reg.predict(np.array([[3, 5]]))
1.0
array([1., 2.])
3.0
array([16.])
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn import metrics
from sklearn.metrics import r2_score
dataset=pd.read_csv('Salary_Data.csv')
dataset.head()
| YearsExperience | Salary | |
|---|---|---|
| 0 | 1.1 | 39343.0 |
| 1 | 1.3 | 46205.0 |
| 2 | 1.5 | 37731.0 |
| 3 | 2.0 | 43525.0 |
| 4 | 2.2 | 39891.0 |
dataset.shape
(30, 2)
dataset
| YearsExperience | Salary | |
|---|---|---|
| 0 | 1.1 | 39343.0 |
| 1 | 1.3 | 46205.0 |
| 2 | 1.5 | 37731.0 |
| 3 | 2.0 | 43525.0 |
| 4 | 2.2 | 39891.0 |
| 5 | 2.9 | 56642.0 |
| 6 | 3.0 | 60150.0 |
| 7 | 3.2 | 54445.0 |
| 8 | 3.2 | 64445.0 |
| 9 | 3.7 | 57189.0 |
| 10 | 3.9 | 63218.0 |
| 11 | 4.0 | 55794.0 |
| 12 | 4.0 | 56957.0 |
| 13 | 4.1 | 57081.0 |
| 14 | 4.5 | 61111.0 |
| 15 | 4.9 | 67938.0 |
| 16 | 5.1 | 66029.0 |
| 17 | 5.3 | 83088.0 |
| 18 | 5.9 | 81363.0 |
| 19 | 6.0 | 93940.0 |
| 20 | 6.8 | 91738.0 |
| 21 | 7.1 | 98273.0 |
| 22 | 7.9 | 101302.0 |
| 23 | 8.2 | 113812.0 |
| 24 | 8.7 | 109431.0 |
| 25 | 9.0 | 105582.0 |
| 26 | 9.5 | 116969.0 |
| 27 | 9.6 | 112635.0 |
| 28 | 10.3 | 122391.0 |
| 29 | 10.5 | 121872.0 |
X = dataset.iloc[:, :-1].values
y = dataset.iloc[:, 1].values
X
y
array([[ 1.1],
[ 1.3],
[ 1.5],
[ 2. ],
[ 2.2],
[ 2.9],
[ 3. ],
[ 3.2],
[ 3.2],
[ 3.7],
[ 3.9],
[ 4. ],
[ 4. ],
[ 4.1],
[ 4.5],
[ 4.9],
[ 5.1],
[ 5.3],
[ 5.9],
[ 6. ],
[ 6.8],
[ 7.1],
[ 7.9],
[ 8.2],
[ 8.7],
[ 9. ],
[ 9.5],
[ 9.6],
[10.3],
[10.5]])
array([ 39343., 46205., 37731., 43525., 39891., 56642., 60150.,
54445., 64445., 57189., 63218., 55794., 56957., 57081.,
61111., 67938., 66029., 83088., 81363., 93940., 91738.,
98273., 101302., 113812., 109431., 105582., 116969., 112635.,
122391., 121872.])
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test=train_test_split(X, y, test_size=0.20)
len(X_test)
6
from sklearn.linear_model import LinearRegression
regressor=LinearRegression()
regressor.fit(X_train,y_train)
LinearRegression()
y_pred=regressor.predict(X_test)
y_pred
y_test
np.sqrt(metrics.mean_squared_error(y_test, y_pred))
array([ 82018.60003626, 40458.31505268, 72573.08072181, 123578.88501985,
36680.1073269 , 68794.87299603])
array([ 81363., 37731., 67938., 122391., 39343., 61111.])
4018.6234071254835
r2_score(y_test, y_pred)
0.9803107390583423
regressor.coef_
array([9445.51931445])
regressor.intercept_
26290.036080998558
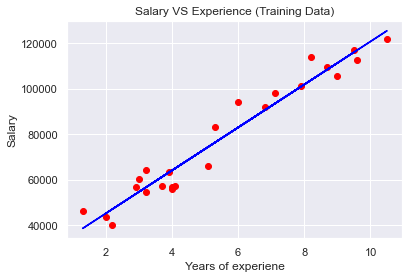
plt.scatter(X_train,y_train,color='red')
plt.plot(X_train,regressor.predict(X_train),color='blue')
plt.title('Salary VS Experience (Training Data)')
plt.xlabel('Years of experiene')
plt.ylabel('Salary')
plt.show()
<matplotlib.collections.PathCollection at 0x170fab89670>
[<matplotlib.lines.Line2D at 0x170fab90700>]
Text(0.5, 1.0, 'Salary VS Experience (Training Data)')
Text(0.5, 0, 'Years of experiene')
Text(0, 0.5, 'Salary')
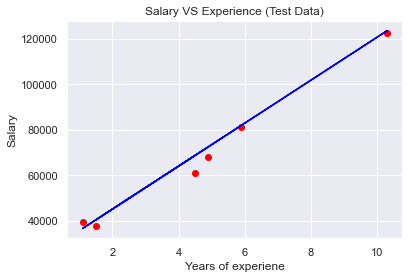
plt.scatter(X_test,y_test,color='red')
plt.plot(X_test,regressor.predict(X_test),color='blue')
plt.title('Salary VS Experience (Test Data)');
plt.xlabel('Years of experiene');
plt.ylabel('Salary');
plt.show()
<matplotlib.collections.PathCollection at 0x170fab2fa60>
[<matplotlib.lines.Line2D at 0x170fab25ee0>]
Text(0.5, 1.0, 'Salary VS Experience (Test Data)')
Text(0.5, 0, 'Years of experiene')
Text(0, 0.5, 'Salary')
Naive Bayes#
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.naive_bayes import GaussianNB
# Load the data
from sklearn.datasets import load_iris
iris = load_iris()
from matplotlib import pyplot as plt
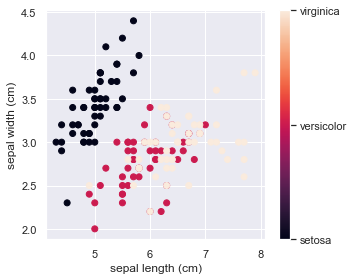
# The indices of the features that we are plotting
x_index = 0
y_index = 1
# this formatter will label the colorbar with the correct target names
formatter = plt.FuncFormatter(lambda i, *args: iris.target_names[int(i)])
plt.figure(figsize=(5, 4))
plt.scatter(iris.data[:, x_index], iris.data[:, y_index], c=iris.target)
plt.colorbar(ticks=[0, 1, 2], format=formatter)
plt.xlabel(iris.feature_names[x_index])
plt.ylabel(iris.feature_names[y_index])
plt.tight_layout()
plt.show()
<Figure size 360x288 with 0 Axes>
<matplotlib.collections.PathCollection at 0x170fab46c10>
<matplotlib.colorbar.Colorbar at 0x170faa3f880>
Text(0.5, 0, 'sepal length (cm)')
Text(0, 0.5, 'sepal width (cm)')
irisdata = load_iris()
print(irisdata.DESCR)
.. _iris_dataset:
Iris plants dataset
--------------------
**Data Set Characteristics:**
:Number of Instances: 150 (50 in each of three classes)
:Number of Attributes: 4 numeric, predictive attributes and the class
:Attribute Information:
- sepal length in cm
- sepal width in cm
- petal length in cm
- petal width in cm
- class:
- Iris-Setosa
- Iris-Versicolour
- Iris-Virginica
:Summary Statistics:
============== ==== ==== ======= ===== ====================
Min Max Mean SD Class Correlation
============== ==== ==== ======= ===== ====================
sepal length: 4.3 7.9 5.84 0.83 0.7826
sepal width: 2.0 4.4 3.05 0.43 -0.4194
petal length: 1.0 6.9 3.76 1.76 0.9490 (high!)
petal width: 0.1 2.5 1.20 0.76 0.9565 (high!)
============== ==== ==== ======= ===== ====================
:Missing Attribute Values: None
:Class Distribution: 33.3% for each of 3 classes.
:Creator: R.A. Fisher
:Donor: Michael Marshall (MARSHALL%PLU@io.arc.nasa.gov)
:Date: July, 1988
The famous Iris database, first used by Sir R.A. Fisher. The dataset is taken
from Fisher's paper. Note that it's the same as in R, but not as in the UCI
Machine Learning Repository, which has two wrong data points.
This is perhaps the best known database to be found in the
pattern recognition literature. Fisher's paper is a classic in the field and
is referenced frequently to this day. (See Duda & Hart, for example.) The
data set contains 3 classes of 50 instances each, where each class refers to a
type of iris plant. One class is linearly separable from the other 2; the
latter are NOT linearly separable from each other.
.. topic:: References
- Fisher, R.A. "The use of multiple measurements in taxonomic problems"
Annual Eugenics, 7, Part II, 179-188 (1936); also in "Contributions to
Mathematical Statistics" (John Wiley, NY, 1950).
- Duda, R.O., & Hart, P.E. (1973) Pattern Classification and Scene Analysis.
(Q327.D83) John Wiley & Sons. ISBN 0-471-22361-1. See page 218.
- Dasarathy, B.V. (1980) "Nosing Around the Neighborhood: A New System
Structure and Classification Rule for Recognition in Partially Exposed
Environments". IEEE Transactions on Pattern Analysis and Machine
Intelligence, Vol. PAMI-2, No. 1, 67-71.
- Gates, G.W. (1972) "The Reduced Nearest Neighbor Rule". IEEE Transactions
on Information Theory, May 1972, 431-433.
- See also: 1988 MLC Proceedings, 54-64. Cheeseman et al"s AUTOCLASS II
conceptual clustering system finds 3 classes in the data.
- Many, many more ...
irisdata.data
array([[5.1, 3.5, 1.4, 0.2],
[4.9, 3. , 1.4, 0.2],
[4.7, 3.2, 1.3, 0.2],
[4.6, 3.1, 1.5, 0.2],
[5. , 3.6, 1.4, 0.2],
[5.4, 3.9, 1.7, 0.4],
[4.6, 3.4, 1.4, 0.3],
[5. , 3.4, 1.5, 0.2],
[4.4, 2.9, 1.4, 0.2],
[4.9, 3.1, 1.5, 0.1],
[5.4, 3.7, 1.5, 0.2],
[4.8, 3.4, 1.6, 0.2],
[4.8, 3. , 1.4, 0.1],
[4.3, 3. , 1.1, 0.1],
[5.8, 4. , 1.2, 0.2],
[5.7, 4.4, 1.5, 0.4],
[5.4, 3.9, 1.3, 0.4],
[5.1, 3.5, 1.4, 0.3],
[5.7, 3.8, 1.7, 0.3],
[5.1, 3.8, 1.5, 0.3],
[5.4, 3.4, 1.7, 0.2],
[5.1, 3.7, 1.5, 0.4],
[4.6, 3.6, 1. , 0.2],
[5.1, 3.3, 1.7, 0.5],
[4.8, 3.4, 1.9, 0.2],
[5. , 3. , 1.6, 0.2],
[5. , 3.4, 1.6, 0.4],
[5.2, 3.5, 1.5, 0.2],
[5.2, 3.4, 1.4, 0.2],
[4.7, 3.2, 1.6, 0.2],
[4.8, 3.1, 1.6, 0.2],
[5.4, 3.4, 1.5, 0.4],
[5.2, 4.1, 1.5, 0.1],
[5.5, 4.2, 1.4, 0.2],
[4.9, 3.1, 1.5, 0.2],
[5. , 3.2, 1.2, 0.2],
[5.5, 3.5, 1.3, 0.2],
[4.9, 3.6, 1.4, 0.1],
[4.4, 3. , 1.3, 0.2],
[5.1, 3.4, 1.5, 0.2],
[5. , 3.5, 1.3, 0.3],
[4.5, 2.3, 1.3, 0.3],
[4.4, 3.2, 1.3, 0.2],
[5. , 3.5, 1.6, 0.6],
[5.1, 3.8, 1.9, 0.4],
[4.8, 3. , 1.4, 0.3],
[5.1, 3.8, 1.6, 0.2],
[4.6, 3.2, 1.4, 0.2],
[5.3, 3.7, 1.5, 0.2],
[5. , 3.3, 1.4, 0.2],
[7. , 3.2, 4.7, 1.4],
[6.4, 3.2, 4.5, 1.5],
[6.9, 3.1, 4.9, 1.5],
[5.5, 2.3, 4. , 1.3],
[6.5, 2.8, 4.6, 1.5],
[5.7, 2.8, 4.5, 1.3],
[6.3, 3.3, 4.7, 1.6],
[4.9, 2.4, 3.3, 1. ],
[6.6, 2.9, 4.6, 1.3],
[5.2, 2.7, 3.9, 1.4],
[5. , 2. , 3.5, 1. ],
[5.9, 3. , 4.2, 1.5],
[6. , 2.2, 4. , 1. ],
[6.1, 2.9, 4.7, 1.4],
[5.6, 2.9, 3.6, 1.3],
[6.7, 3.1, 4.4, 1.4],
[5.6, 3. , 4.5, 1.5],
[5.8, 2.7, 4.1, 1. ],
[6.2, 2.2, 4.5, 1.5],
[5.6, 2.5, 3.9, 1.1],
[5.9, 3.2, 4.8, 1.8],
[6.1, 2.8, 4. , 1.3],
[6.3, 2.5, 4.9, 1.5],
[6.1, 2.8, 4.7, 1.2],
[6.4, 2.9, 4.3, 1.3],
[6.6, 3. , 4.4, 1.4],
[6.8, 2.8, 4.8, 1.4],
[6.7, 3. , 5. , 1.7],
[6. , 2.9, 4.5, 1.5],
[5.7, 2.6, 3.5, 1. ],
[5.5, 2.4, 3.8, 1.1],
[5.5, 2.4, 3.7, 1. ],
[5.8, 2.7, 3.9, 1.2],
[6. , 2.7, 5.1, 1.6],
[5.4, 3. , 4.5, 1.5],
[6. , 3.4, 4.5, 1.6],
[6.7, 3.1, 4.7, 1.5],
[6.3, 2.3, 4.4, 1.3],
[5.6, 3. , 4.1, 1.3],
[5.5, 2.5, 4. , 1.3],
[5.5, 2.6, 4.4, 1.2],
[6.1, 3. , 4.6, 1.4],
[5.8, 2.6, 4. , 1.2],
[5. , 2.3, 3.3, 1. ],
[5.6, 2.7, 4.2, 1.3],
[5.7, 3. , 4.2, 1.2],
[5.7, 2.9, 4.2, 1.3],
[6.2, 2.9, 4.3, 1.3],
[5.1, 2.5, 3. , 1.1],
[5.7, 2.8, 4.1, 1.3],
[6.3, 3.3, 6. , 2.5],
[5.8, 2.7, 5.1, 1.9],
[7.1, 3. , 5.9, 2.1],
[6.3, 2.9, 5.6, 1.8],
[6.5, 3. , 5.8, 2.2],
[7.6, 3. , 6.6, 2.1],
[4.9, 2.5, 4.5, 1.7],
[7.3, 2.9, 6.3, 1.8],
[6.7, 2.5, 5.8, 1.8],
[7.2, 3.6, 6.1, 2.5],
[6.5, 3.2, 5.1, 2. ],
[6.4, 2.7, 5.3, 1.9],
[6.8, 3. , 5.5, 2.1],
[5.7, 2.5, 5. , 2. ],
[5.8, 2.8, 5.1, 2.4],
[6.4, 3.2, 5.3, 2.3],
[6.5, 3. , 5.5, 1.8],
[7.7, 3.8, 6.7, 2.2],
[7.7, 2.6, 6.9, 2.3],
[6. , 2.2, 5. , 1.5],
[6.9, 3.2, 5.7, 2.3],
[5.6, 2.8, 4.9, 2. ],
[7.7, 2.8, 6.7, 2. ],
[6.3, 2.7, 4.9, 1.8],
[6.7, 3.3, 5.7, 2.1],
[7.2, 3.2, 6. , 1.8],
[6.2, 2.8, 4.8, 1.8],
[6.1, 3. , 4.9, 1.8],
[6.4, 2.8, 5.6, 2.1],
[7.2, 3. , 5.8, 1.6],
[7.4, 2.8, 6.1, 1.9],
[7.9, 3.8, 6.4, 2. ],
[6.4, 2.8, 5.6, 2.2],
[6.3, 2.8, 5.1, 1.5],
[6.1, 2.6, 5.6, 1.4],
[7.7, 3. , 6.1, 2.3],
[6.3, 3.4, 5.6, 2.4],
[6.4, 3.1, 5.5, 1.8],
[6. , 3. , 4.8, 1.8],
[6.9, 3.1, 5.4, 2.1],
[6.7, 3.1, 5.6, 2.4],
[6.9, 3.1, 5.1, 2.3],
[5.8, 2.7, 5.1, 1.9],
[6.8, 3.2, 5.9, 2.3],
[6.7, 3.3, 5.7, 2.5],
[6.7, 3. , 5.2, 2.3],
[6.3, 2.5, 5. , 1.9],
[6.5, 3. , 5.2, 2. ],
[6.2, 3.4, 5.4, 2.3],
[5.9, 3. , 5.1, 1.8]])
irisdata.target
array([0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2,
2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2, 2])
X = irisdata.data
Y = irisdata.target
X_train, X_test, y_train, y_test=train_test_split(X, Y, test_size=0.2, random_state=0)
gnb = GaussianNB()
gnb.fit(X_train, y_train)
GaussianNB()
y_pred = gnb.predict(X_test)
100*metrics.accuracy_score(y_test, y_pred)
96.66666666666667
# 400
380 # disease free
20 # disease
380/400
380
20
0.95
y_test
array([2, 1, 0, 2, 0, 2, 0, 1, 1, 1, 2, 1, 1, 1, 1, 0, 1, 1, 0, 0, 2, 1,
0, 0, 2, 0, 0, 1, 1, 0])
y_pred
array([2, 1, 0, 2, 0, 2, 0, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 1, 0, 0, 2, 1,
0, 0, 2, 0, 0, 1, 1, 0])

